Exploring chromatin accessibility and its diagnostic potential in cancer through the utilization of cell-free DNA
This study is led by Dr. Xun Lan (Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing, China), Dr. Jiafu Ji, Dr. Zhaode Bu (Center of Gastrointestinal Cancer, Peking University Cancer Hospital & Institute, Beijing, China), Dr. Honghao Yu (Guangxi Key Laboratory of Drug Discovery and Optimization, Guilin medical University), Dr. Wenming Wu, Dr. Yupei Zhao, Dr. Junya Peng (Peking Union Medical College Hospital, Peking Union Medical College, Beijing) and Dr. Peilin Cui (Tiantan Hospital of Capital Medical University, Beijing, China).
Credit: ©Science China Press
This study is led by Dr. Xun Lan (Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing, China), Dr. Jiafu Ji, Dr. Zhaode Bu (Center of Gastrointestinal Cancer, Peking University Cancer Hospital & Institute, Beijing, China), Dr. Honghao Yu (Guangxi Key Laboratory of Drug Discovery and Optimization, Guilin medical University), Dr. Wenming Wu, Dr. Yupei Zhao, Dr. Junya Peng (Peking Union Medical College Hospital, Peking Union Medical College, Beijing) and Dr. Peilin Cui (Tiantan Hospital of Capital Medical University, Beijing, China).
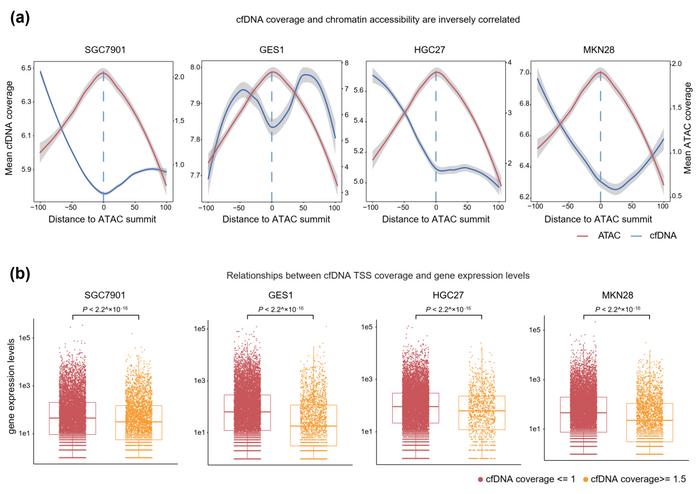
Deciphering tumor cell gene expression patterns can be deduced from circulating cell-free DNA (cfDNA) characteristics, such as histone modifications and fragmentation patterns at promoters. However, the direct correlation between cfDNA features and tumor-specific chromatin accessibility remains to be conclusively established, thereby restricting its current application primarily to cancer diagnosis. In genes undergoing active transcription, promoters experience nucleosome dissociation, diminishing their preservation and detectability within cfDNA. Consequently, the extent of cfDNA coverage at promoter regions can serve as a proxy for the chromatin landscape, reflecting gene expression levels within viable cells.
In vitro cell-culture cfDNA model unveiled that genes exhibiting low coverage of cfDNA transcript start sites (TSS) were associated with heightened chromatin accessibility and elevated expression levels. they performed ATAC-seq and RNA-seq analyses on four cell lines (SGC7901, GES1, HGC27, and MKN28). Additionally, sequencing analysis was conducted on cfDNA in the extracellular culture medium (DMEM). The study indicated that the TSS coverage of cfDNA is negatively correlated with the chromatin accessibility and transcription levels of genes.”This cell line experiment is very interesting,” Xun says.
In addition, the cfDNA TSS coverage in blood samples from STAD and BRCA patients showed a negative association with ATAC-seq coverage in the corresponding tumor tissues from the TCGA dataset. The Pearson correlation coefficient was -0.65 in STAD (P value < 2.2×10-16) and -0.55 in BRCA (P value < 2.2×10-16). Moreover, genes with high ATAC-seq signals in tumor tissues exhibited lower cfDNA TSS coverage compared to genes with low cfDNA TSS coverage (P value < 2.2×10-16 for both STAD and BRCA).
Regarding the TCGA STAD RNA-seq datasets, genes with high cfDNA TSS coverage demonstrated significantly lower expression levels compared to genes with low cfDNA TSS coverage (P value < 2.2×10-16). Genes with low cfDNA TSS coverage were enriched in various cancer-related pathways and biological functions, including gastric cancer, the cell cycle, the MAPK signaling pathway, and the Wnt signaling pathway. Next, they calculated the differentially expressed patterns of all genes between tumor and normal tissues from the TCGA STAD RNA-seq data and TCGA BRCA RNA-seq data. Consistent with the findings from in vitro cell lines, upregulated genes in TCGA STAD tumor samples and TCGA BRCA tumor samples exhibited significantly lower cfDNA TSS coverage compared to downregulated genes (P value < 2.2×10-16 in cfDNA TSS coverage).
Moreover, The team conducted an exhaustive analysis of plasma cfDNA sourced from 546 individuals, encompassing patients with 21 diverse disease types as well as healthy donors. They found that this inverse relationship could identify patients with cancer. They collected peripheral blood cfDNA from a broad cohort, including patients with gastric cancer (n=101), breast cancer (n=57), pancreatic cancer (n=40), gastritis (n=83), non-cancer diseases (such as bone disease, cerebral hemorrhage, heart disease, benign tumors, etc.) (n=58), and healthy individuals (n=100) to study cancer-specific cfDNA characteristics. The study found that cfDNA coverage in promoter regions could accurately distinguish gastric cancer (STAD, AUC 0.96), breast cancer (BRCA, AUC 0.98), and pancreatic ductal adenocarcinoma (PDAC, AUC 0.88) from healthy individuals, as well as from patients with gastritis, non-cancer diseases, and other types of cancer.
They further explored the cfDNA TSS coverage of these cancer-type-specific genes in an independent cohort. They developed a targeted sequencing assay called cfDNA Open Chromatin Capture Sequencing (CFOCUS) using hybridization capture of the STAD-specific genes and validated its performance in a cohort of 80 plasma samples, including 35 gastric cancer patients and 45 healthy individuals. These 35 patients with gastric cancer included early-stage samples (stage I and stage II), which strengthened the robustness of cancer screening. Using the same model built for the WGS data of cfDNA from patients with STAD, CFOCUS samples with a cancer risk proportion greater than 0.5 were considered as patients with STAD. CFOCUS achieved an AUC of 0.88 (80% sensitivity and 91% specificity). The mean Pearson coefficient among three repeated cfDNA samples for each healthy donor was 0.93, 0.88, and 0.90, underscoring the robustness of the custom probe panel utilized in their study. These results demonstrate the potential utility of cfDNA coverage features in early cancer detection.
See the article:
Uncovering chromatin accessibility and cancer diagnostic potential via cell-free DNA utilization
Journal
Science Bulletin
DOI
10.1016/j.scib.2024.04.013